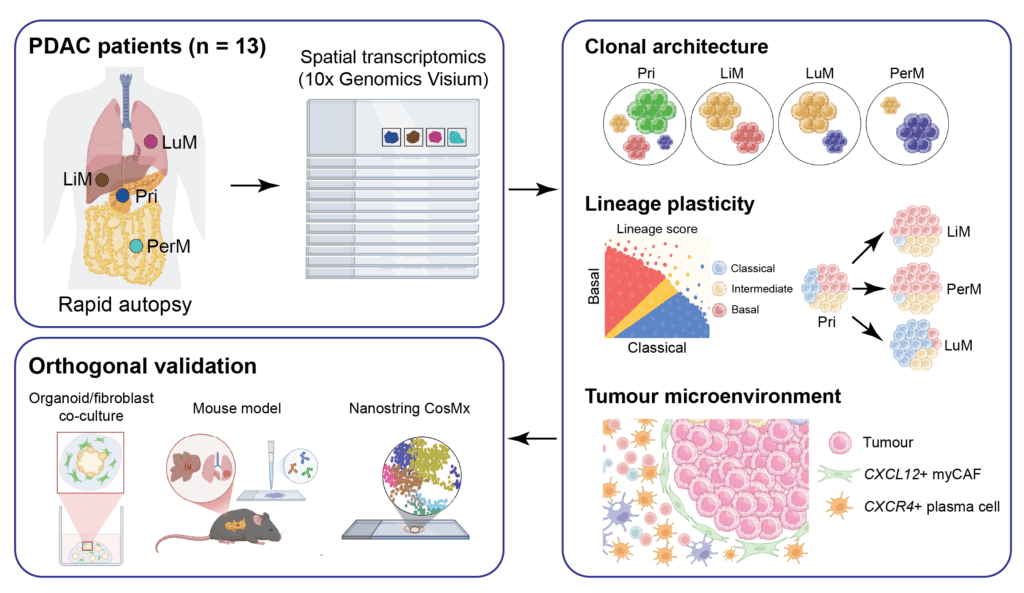
Pancreatic cancer remains one of the most lethal malignancies, largely due to its propensity for systemic dissemination and its profound cellular heterogeneity. Despite these challenges, the mechanisms by which tumour cells adapt to diverse metastatic niches and evade immune surveillance have remained elusive, particularly from a spatial perspective. In this study, we applied spatial transcriptomics to a uniquely valuable cohort of primary tumours and multi-organ metastases obtained through rapid autopsy. By mapping clonal architecture, lineage plasticity, and tumour microenvironmental features, we delineated striking site-specific transcriptional adaptations, including pronounced intra-patient distinctions between liver and lung metastases. Notably, even within the same organ – most prominently in the liver – clonal architecture and lineage plasticity often diverged across metastatic foci, underscoring the remarkable heterogeneity of tumour dissemination. Importantly, the study identified a robust association between basal-like tumour states and TGFB1-expressing myofibroblastic cancer-associated fibroblasts (myCAFs), which orchestrate plasma-cell exclusion through CXCR4–CXCL12 signaling. Validation across patient-derived organoids and cross-species models further underscored the dynamic crosstalk between tumour lineages and their stromal ecosystems. These insights not only reveal the profound spatial heterogeneity underlying therapy-refractory pancreatic cancer but also highlight stromal-tumour interactions as actionable vulnerabilities for future therapeutic development.
.
 Summary of the findings
Summary of the findings